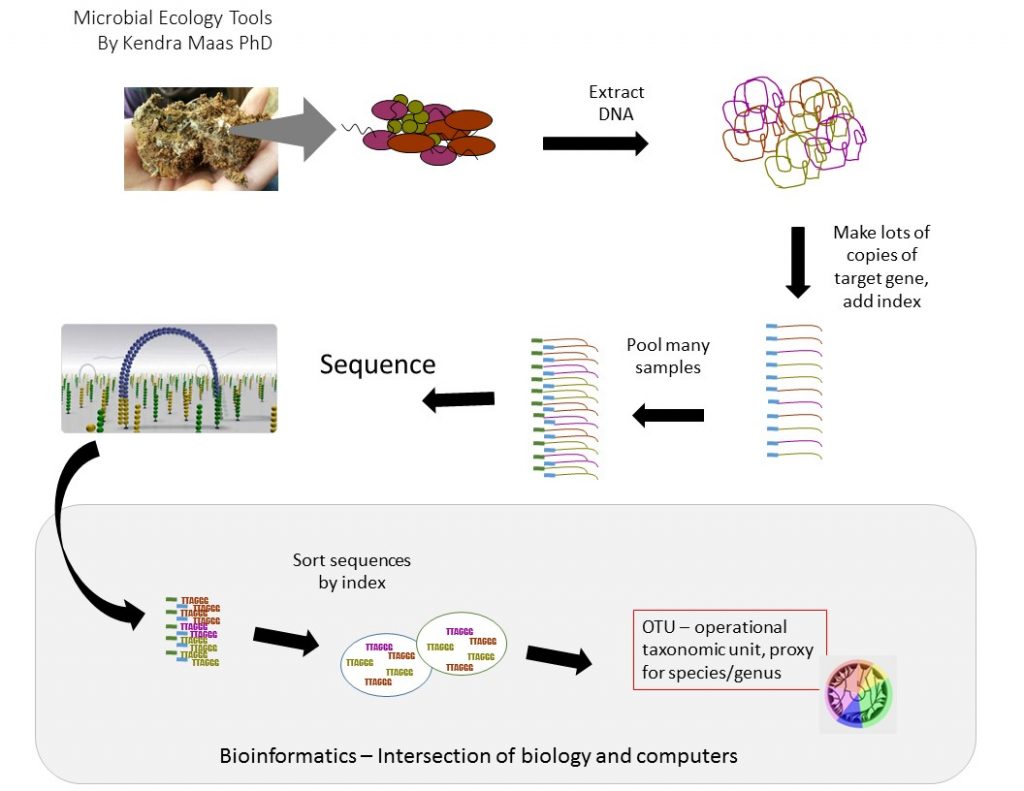
Microbiome and Amplicon Sequencing
Microbial Ecology has been revolutionized by DNA sequencing. We leverage the low per base cost of Illumina next gen sequencing and deep multiplexing (barcoding) to keep our per sample costs low. We offer both project or “fill the run” based sequencing. “Fill the run” means that we don’t have sample minimums, we will process samples from smaller projects together to keep the costs as low as possible. CGI offers several on-ramps for this analysis. Full service sample processing is available by combining DNA extraction with our amplicon library prep and amplicon sequencing which includes quantification, library prep, and sequencing at least 10,000 sequences per sample. We have custom, dual indexed library prepration primers for Bacterial v4 16S (HMP 515F/806R), Bacterial and Archaeal v4 16S (EMP 515F/806R), Fungal ITS2 (fITS7/ITS4), and user specified 2 step.